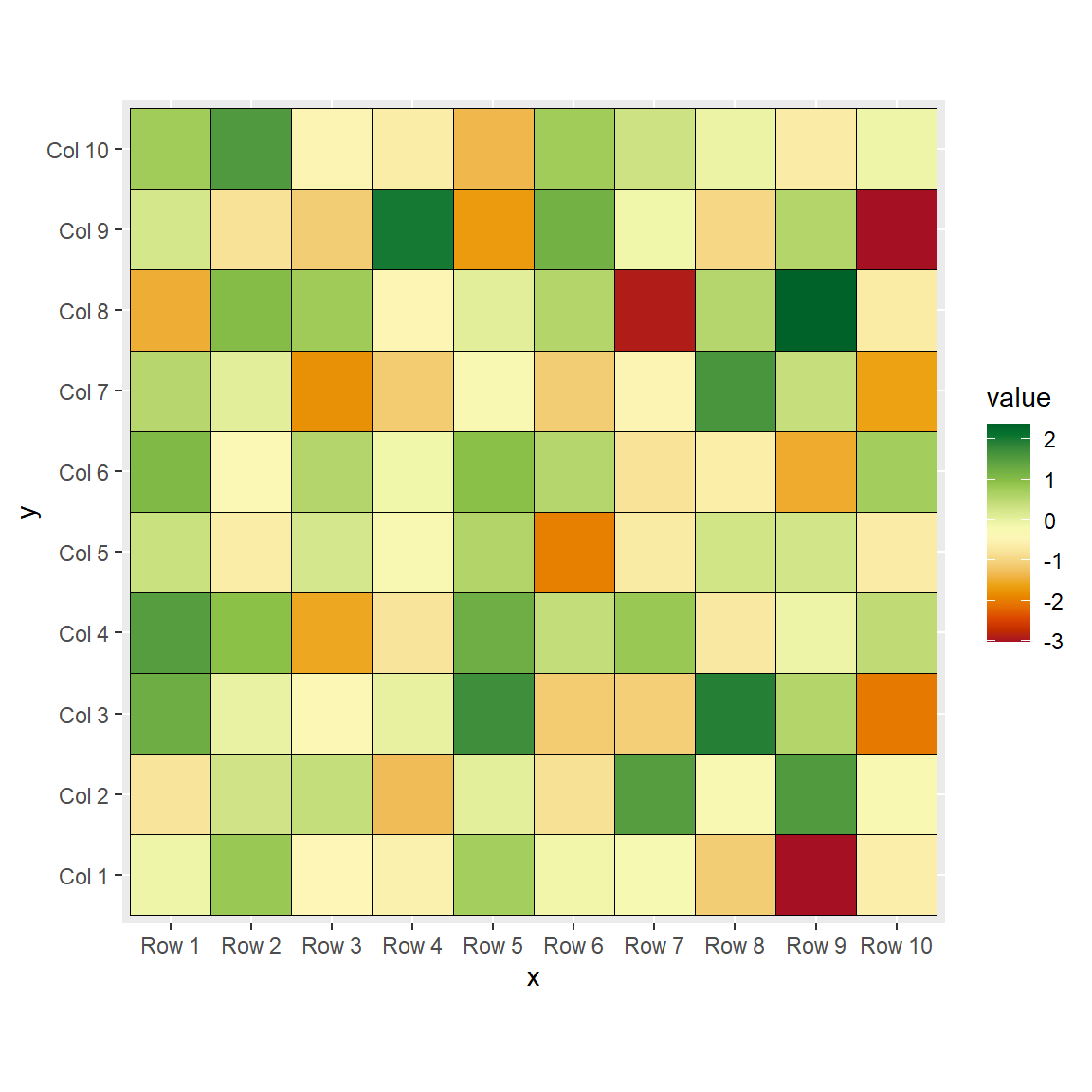
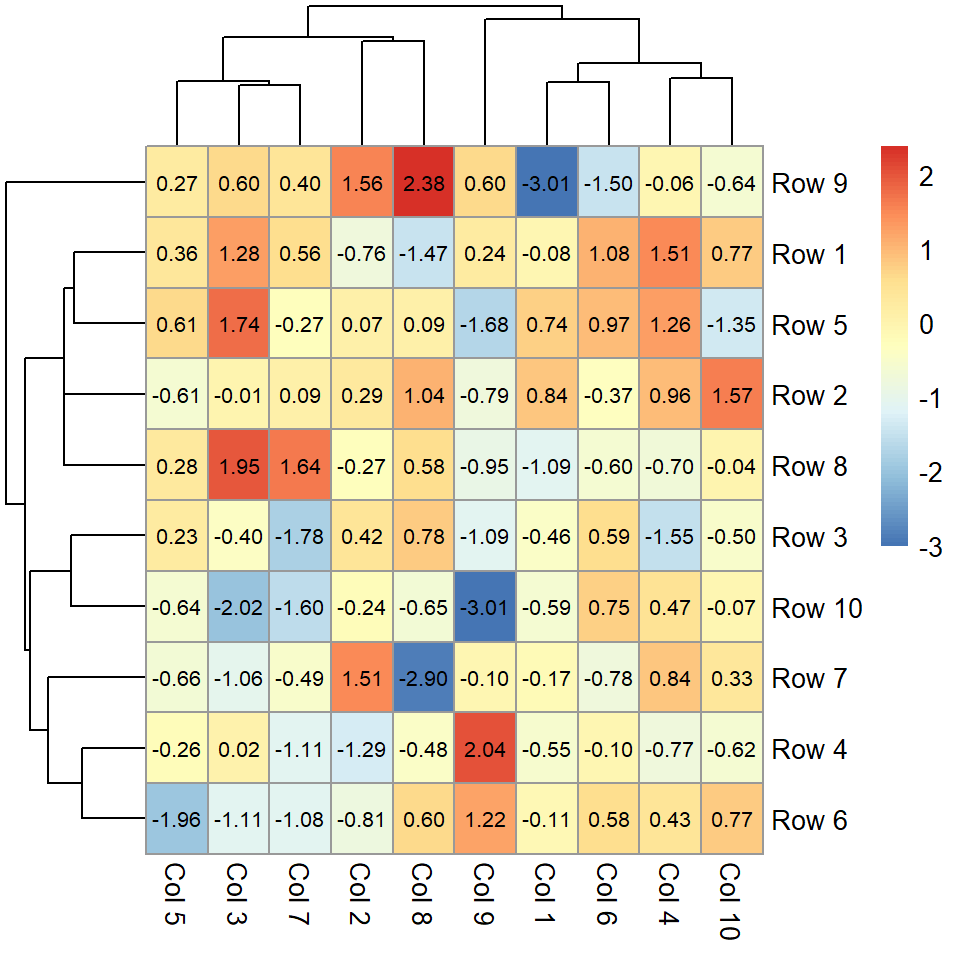
The heatmap function
The heatmap function can draw a heat map in R from a matrix. In the following examples we are going to use a square matrix but note that the number of rows and columns doesn’t need to be the same.
# Matrix
m <- matrix(rnorm(100), ncol = 10)
colnames(m) <- paste("Col", 1:10)
rownames(m) <- paste("Row", 1:10)
heatmap(m)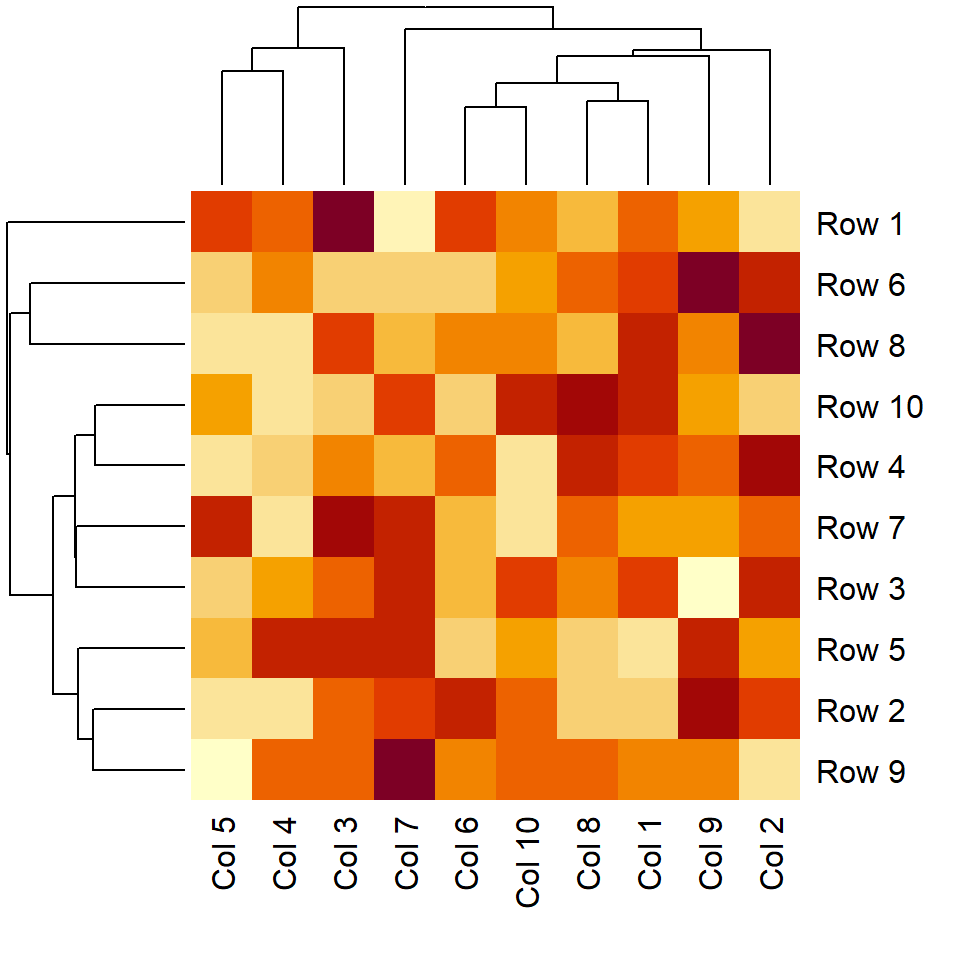
Data normalization
The data is normalized by rows by default. However, you might need to normalize it in the column direction ("column") or if no normalization must be carried at all ("none").
# Matrix
m <- matrix(rnorm(100), ncol = 10)
colnames(m) <- paste("Col", 1:10)
rownames(m) <- paste("Row", 1:10)
heatmap(m, scale = "column")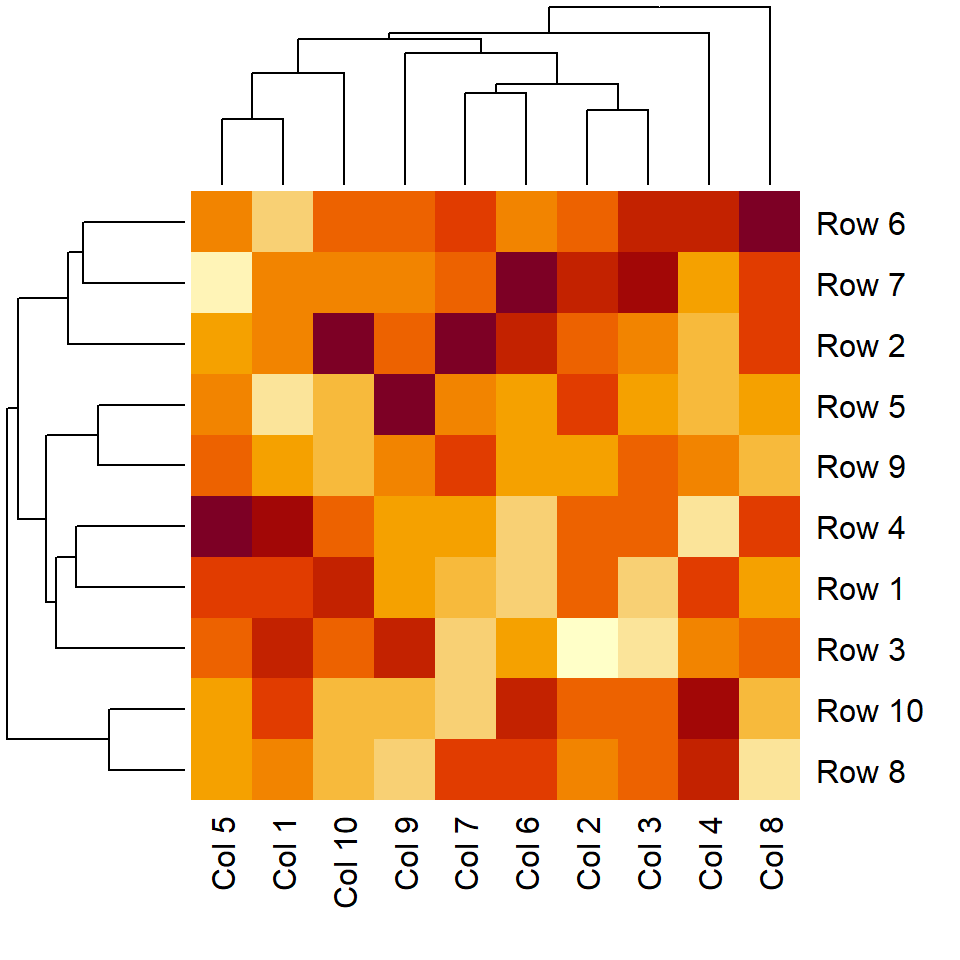
Color customization
You can pass a color palette to the col argument of the heatmap function. In the following example we are using the viridis palette via hcl.colors.
# Matrix
m <- matrix(rnorm(100), ncol = 10)
colnames(m) <- paste("Col", 1:10)
rownames(m) <- paste("Row", 1:10)
heatmap(m, col = hcl.colors(50))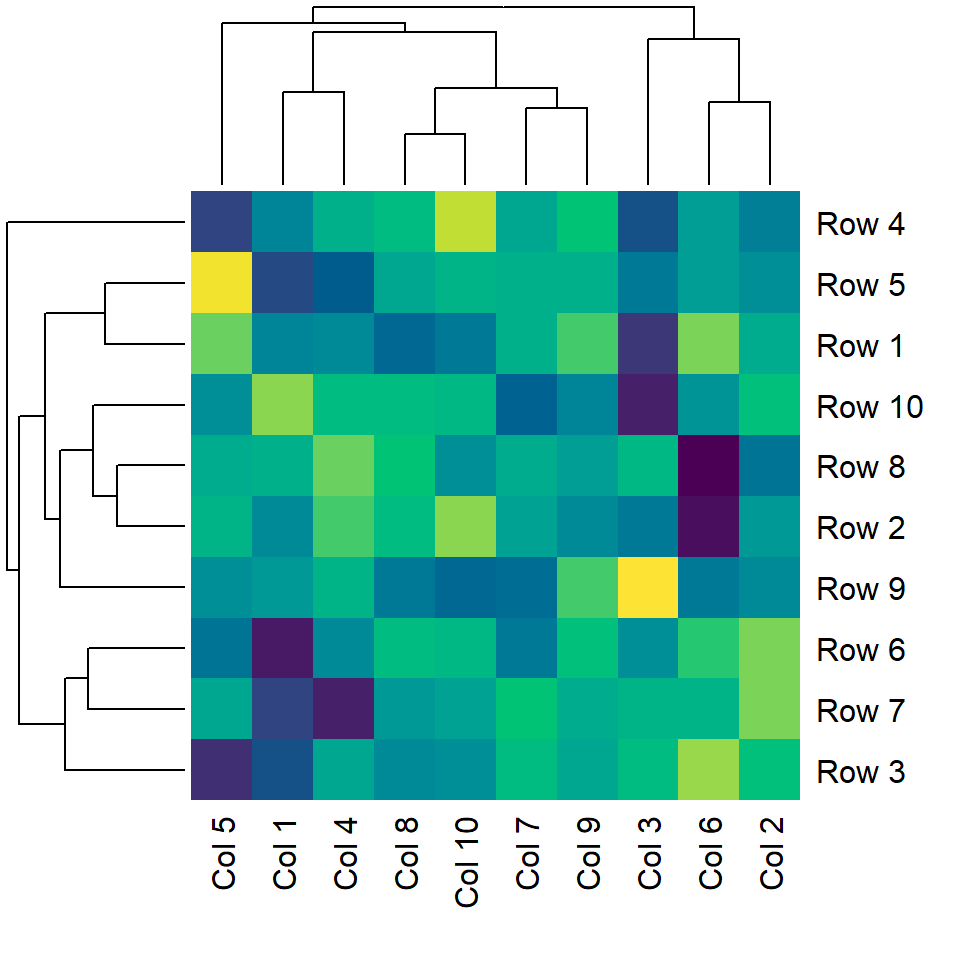
Side colors
It is possible to pass a vector of colors to the ColSideColors and RowSideColors argument to annotate the columns and the rows of the matrix, respectively.
# Matrix
m <- matrix(rnorm(100), ncol = 10)
colnames(m) <- paste("Col", 1:10)
rownames(m) <- paste("Row", 1:10)
heatmap(m,
ColSideColors = rainbow(ncol(m)),
RowSideColors = rainbow(nrow(m)))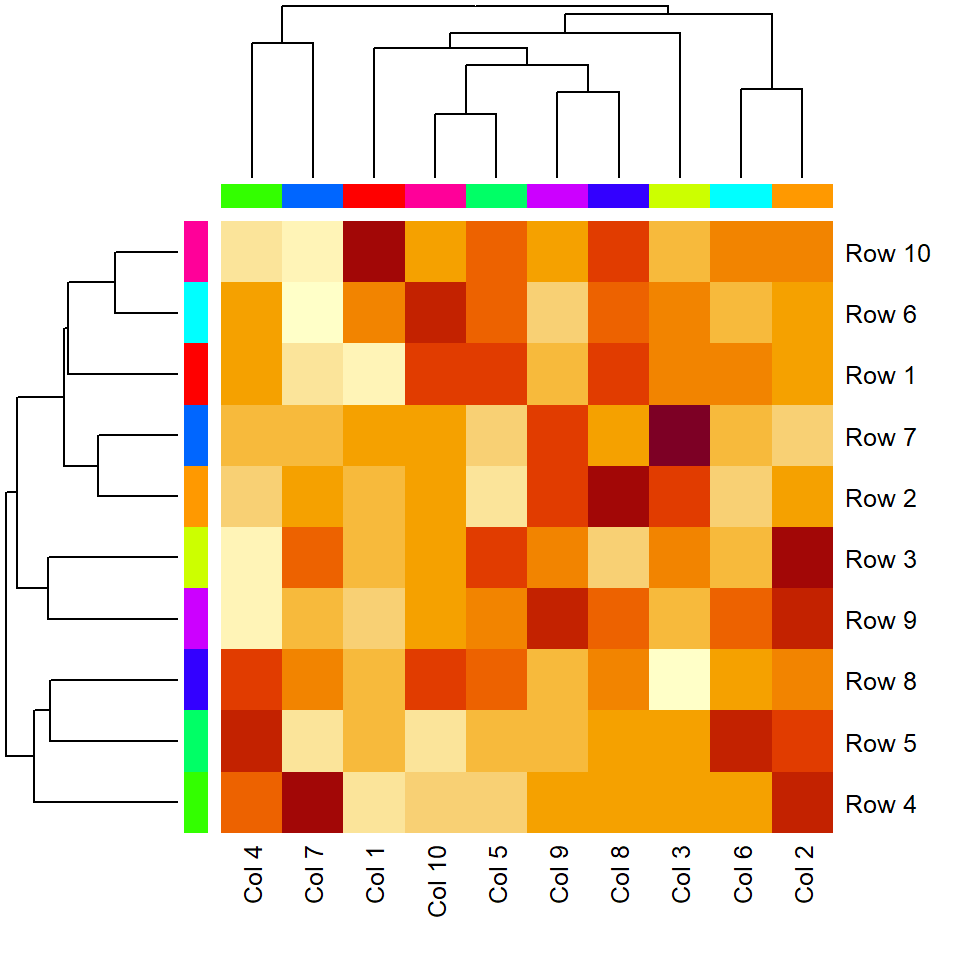
Heat map dendrograms
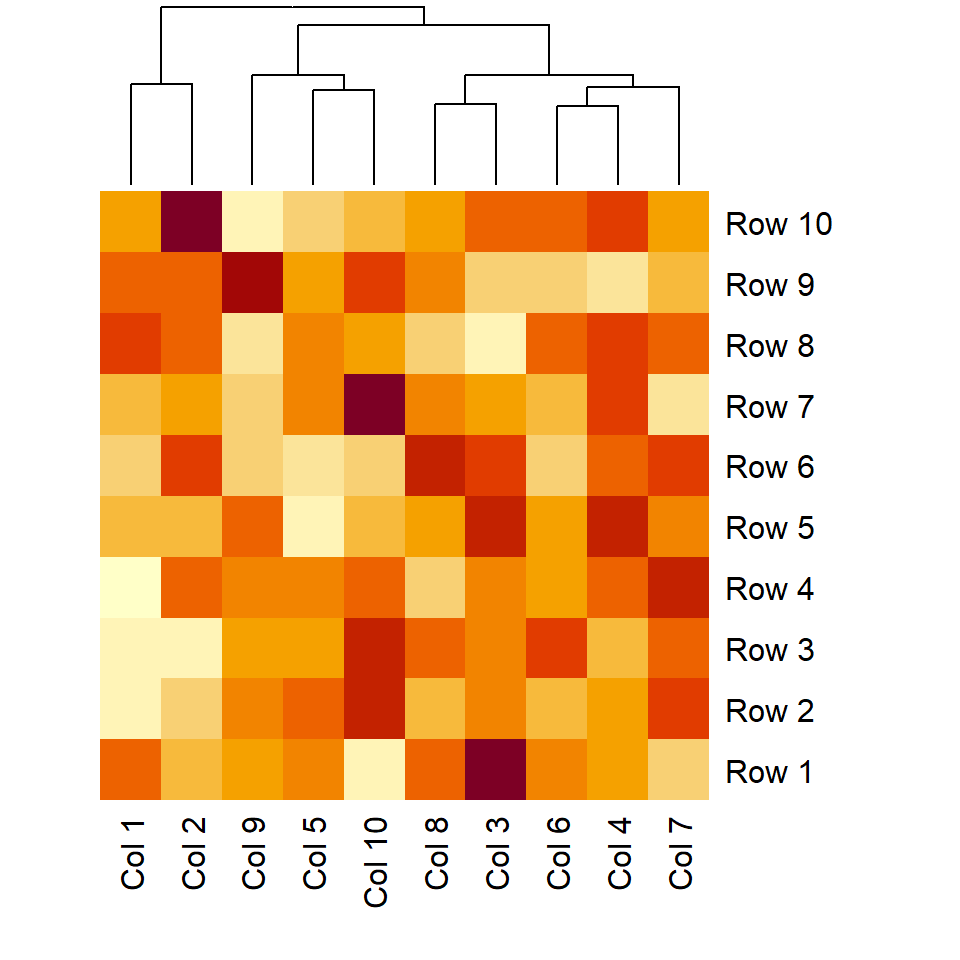
Remove row dendrogram
The Rowv argument controls if the row dendrogram should be computed and how. You can pass a dendrogram or a vector specifying the order. Setting it to NA will remove the dendrogram.
# Matrix
m <- matrix(rnorm(100), ncol = 10)
colnames(m) <- paste("Col", 1:10)
rownames(m) <- paste("Row", 1:10)
heatmap(m, Rowv = NA)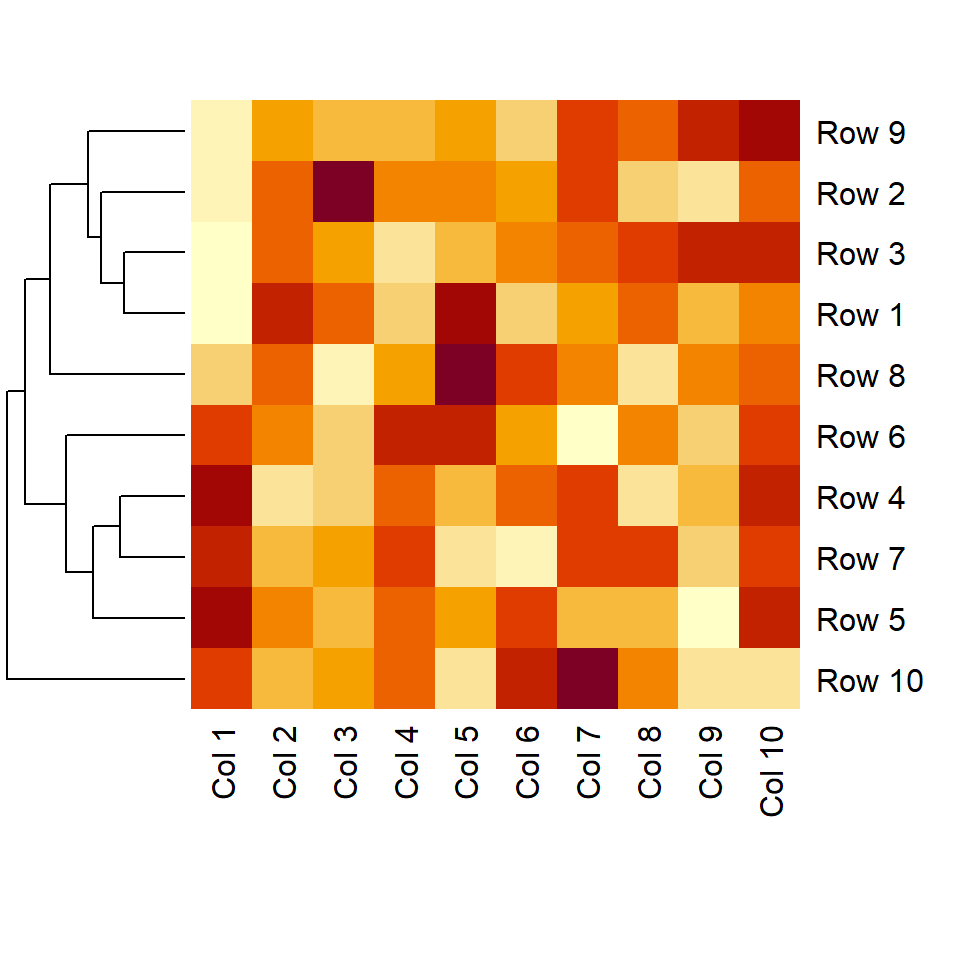
Remove column dendrogram
The Colv argument is the equivalent of Rowv but for columns. Set it to NA to remove the column dendrogram.
# Matrix
m <- matrix(rnorm(100), ncol = 10)
colnames(m) <- paste("Col", 1:10)
rownames(m) <- paste("Row", 1:10)
heatmap(m, Colv = NA)
Remove both dendrograms
Setting Rowv and Colv to NA will remove both dendrograms of the heat map and no reordering will be computed.
# Matrix
m <- matrix(rnorm(100), ncol = 10)
colnames(m) <- paste("Col", 1:10)
rownames(m) <- paste("Row", 1:10)
heatmap(m, Rowv = NA, Colv = NA)